Edinburgh Summer School 2012: tutorial and links, Wednesday 29th August.
New: Stochastic Ion Channel Gating in Dendritic Neurons: Morphology Dependence and Probabilistic Synaptic Activation of Dendritic Spikes in PLoS Computational Biology 6(8): e1000886. doi:10.1371/journal.pcbi.1000886.
Latest changes: psics-1.1.1 (18-May-2011)
PSICS
PSICS computes the behavior of neurons taking account of the stochastic nature of ion channel gating and the detailed positions of the channels themselves. It supports representation of ion channels as kinetic schemes inviolving one or more serial gating complexes.
It uses a variety of approximations to speed up the calculations while retaining effects of channel stochasticity. In particular, it uses population statistics for electrically compact homogeneous channel populations instead of following each channel independently. This contrasts with more costly simulation methods where exact transition times are generated for each stochastic element in the system. As a further approximation, it can compute some or all of the channels in the continuum limit where the behavior is deterministic.
PSICS is intended to be complementary to existing tools. With its focus on kinitec scheme channel models, stochastic behavior, and detailed geometry, it lives in the space between stochastic diffusion models of small sections of neurons (MCell, STEPS) and deterministic whole cell models (Neuron, Genesis).
The user guide explains how to install PSICS, set up models and run them. The examples show input files and results for a range of models and the input formats reference details all the possible options for specifying models.
The best way to hear about the latest developments with PSICS is to register on the forum and switch on notifications for the "Announcements" list.
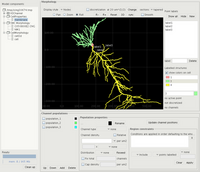
ICING user interface
Models for use with PSICS are expressed as simple XML files (see Rallpack1 for an example) that can be created with an xml-aware text editor. Apart from cell morphologies and ion channels (which rarely need changing) the most complex part of building a model is normally the specification of channel distributions and the positioning of stimuli and recorders. This is the purpose of ICING, a standalone graphical tool for creating and visualizing ion channel distributions.
 |
 |
 |
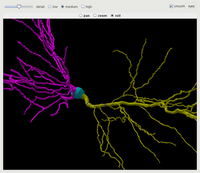
More information on the options for specifying channel densities is in the reference section. Some screenshots show ICING in use and the getting started guide covers the rest.
Features
- Stochastic simulation of ion channels on extended cells
- Spatial discretization (compartmentalization) of cell structures that preserves volume, point positions and surface area
- Kinetic scheme channel models in ensemble and stochastic regimes
- Hodgkin-Huxley style models implemented by automatic conversion to kinetic schemes
- Efficient approximate stochastic simulation methods where the dominant computational cost in many models is the generation of random numbers
- Declarative model specifications in XML
- No manual units matching is required - inputs are specified with their units and converted internally
- Support for standard morphology formats (MorphML, swc, output from Neuron)
- Stand-alone computational core as a native executable for multi-processor applications
- Built-in example models for testing installations and as starting points for new models

