cianmar30 (run.xml)
Channel noise in dendrites
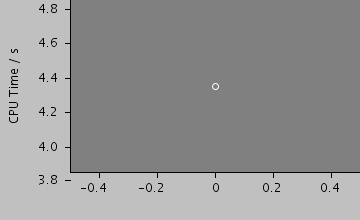
Total CPU time 4.350 seconds; at 12:14:07 Thu 24 Sep 2009
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | Non Gated channels / cpmts | CPU Time / s |
| 1680 | 4978 / 1487 | 0 / 0 | 0 / 0 | 4.35 |
 |
Morphology: ri04NEURON

Predefined views
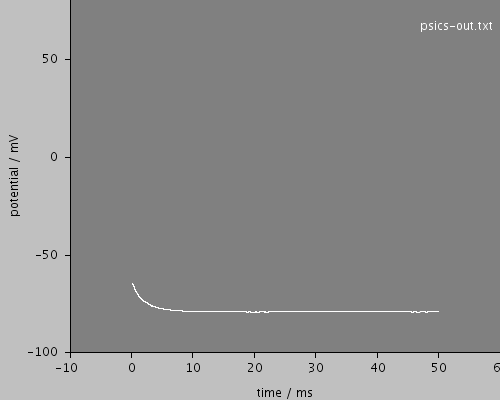
whole
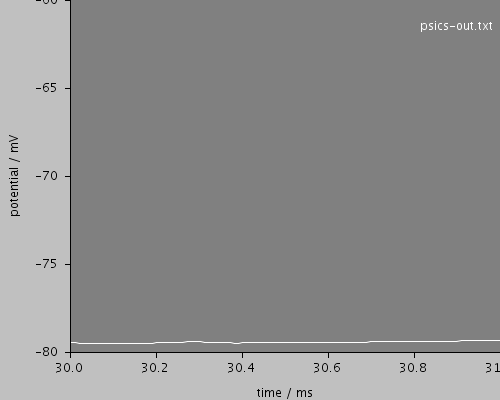
zoom1ms
All files
| Model | Preprocessed | Outupt data | Reference data etc |
| run.xml ri04NEURON.xml membrane.xml environment.xml recording.xml leak-na.xml leak-k.xml |
psics-out.ppp |
log.txt psics-out.sum psics-out.dat psics-out.txt |
Model
Archive file of the complete model: cianmar30.jarrun.xml
<PSICSRun timeStep="0.01ms" runTime="50ms" startPotential="-65mV"
morphology="ri04NEURON"
environment="environment"
properties="membrane"
access="recording"
stochThreshold="1000000"
repeats="1">
<!-- <MorphologySource format="swc" file="ri04.swc"/> -->
<!--<ModelFolder path='../cells'/>-->
<StructureDiscretization baseElementSize="10um"/>
<info>Channel noise in dendrites</info>
<ViewConfig>
<LineGraph width="500" height="400">
<XAxis min="0" max="50" label="time / ms"/>
<YAxis min="-80" max="60" label="potential / mV"/>
<LineSet file="psics-out.txt" color="white"/>
<View id="whole" xmin="-10." xmax="60." ymin="-100." ymax="80."/>
<View id="zoom1ms" xmin="30." xmax="31" ymin="-80." ymax="-60."/>
</LineGraph>
</ViewConfig>
</PSICSRun>
ri04NEURON.xml (truncated)
<?xml version="1.0" encoding="UTF-8"?>
<!--
NEURON version="5.9">
-->
<neuroml xmlns="http://morphml.org/neuroml/schema"
xmlns:mml="http://morphml.org/morphml/schema"
xmlns:meta="http://morphml.org/metadata/schema"
xmlns:bio="http://morphml.org/biophysics/schema"
xmlns:cml="http://morphml.org/channelml/schema"
xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance"
xsi:schemaLocation="http://morphml.org/neuroml/schema ../../Schemata/v1.2/Level2/NeuroML_v1.2.xsd"
name = "Test"
lengthUnits="micron">
<!-- ./ri04NEURON.xml -->
<!-- XML file generated by NEURON 5.9 ModelViewer -->
<!-- Authors: Michael Hines and Sushil Kambampati -->
<!-- Yale University -->
<!-- Date: Sun Apr 13 15:06:23 BST 2008 -->
<morphml>
<cells>
<cell name="soma[0]">
<segments>
<segment id="1" name = "Seg0_soma[0]" cable = "0">
<proximal x="4.4" y="-1.674" z="0" diameter="12.846"/>
<distal x="4.4" y="-0.837" z="0" diameter="12.306"/>
</segment>
<segment id="2" name = "Seg1_soma[0]" parent="1" cable = "0">
<distal x="4.4" y="-0.208" z="0" diameter="12.074"/>
</segment>
<segment id="3" name = "Seg2_soma[0]" parent="2" cable = "0">
<distal x="4.4" y="0.56" z="0" diameter="11.672"/>
</segment>
<segment id="4" name = "Seg3_soma[0]" parent="3" cable = "0">
<distal x="4.4" y="1.141" z="0" diameter="11.478"/>
</segment>
<segment id="5" name = "Seg4_soma[0]" parent="4" cable = "0">
<distal x="4.4" y="1.799" z="0" diameter="11.526"/>
</segment>
<segment id="6" name = "Seg5_soma[0]" parent="5" cable = "0">
<distal x="4.4" y="2.207" z="0" diameter="11.2"/>
</segment>
<segment id="7" name = "Seg6_soma[0]" parent="6" cable = "0">
<distal x="4.4" y="2.831" z="0" diameter="11.036"/>
</segment>
<segment id="8" name = "Seg7_soma[0]" parent="7" cable = "0">
<distal x="4.4" y="3.367" z="0" diameter="11.232"/>
</segment>
<segment id="9" name = "Seg8_soma[0]" parent="8" cable = "0">
<distal x="4.4" y="3.809" z="0" diameter="11.166"/>
</segment>
<segment id="10" name = "Seg9_soma[0]" parent="9" cable = "0">
<distal x="4.4" y="4.499" z="0" diameter="10.194"/>
</segment>
<segment id="11" name = "Seg10_soma[0]" parent="10" cable = "0">
<distal x="4.4" y="4.981" z="0" diameter="10.256"/>
</segment>
<segment id="12" name = "Seg11_soma[0]" parent="11" cable = "0">
<distal x="4.4" y="5.483" z="0" diameter="10.552"/>
</segment>
<segment id="13" name = "Seg12_soma[0]" parent="12" cable = "0">
- and 30094 more lines -membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="150ohm_cm"
membraneCapacitance="0.75uF_per_cm2">
<ChannelPopulation id="leak-na" channel="leak-na" density="0.02per_um2"/>
<ChannelPopulation id="leak-k" channel="leak-k" density="0.22per_um2"/>
</CellProperties>
environment.xml
<CellEnvironment id="environment" temperature="37.0Celsius"> <Ion id="Na" name="Sodium" reversalPotential="50mV"/> <Ion id="K" name="Potassium" reversalPotential="-90mV"/> <Ion id="Ca" name="Calcium" reversalPotential="140mV"/> </CellEnvironment>
recording.xml
<Access id="recording" saveInterval="0.01ms"> <VoltageRecorder at="100"/> </Access>
leak-na.xml
<KSChannel id="leak-na" gSingle="20pS" permeantIon="Na"> <OpenState id="o1"/> <ClosedState id="c1"/> <FixedRateTransition from="o1" to="c1" forward="3.per_ms" reverse="7.per_ms"/> </KSChannel>
leak-k.xml
<KSChannel id="leak-k" gSingle="20pS" permeantIon="K"> <OpenState id="o1"/> <ClosedState id="c1"/> <FixedRateTransition from="o1" to="c1" forward="3.per_ms" reverse="7.per_ms"/> </KSChannel>