mainen (run.xml)
Total CPU time 628.3 seconds; at 12:47:24 Thu 24 Sep 2009
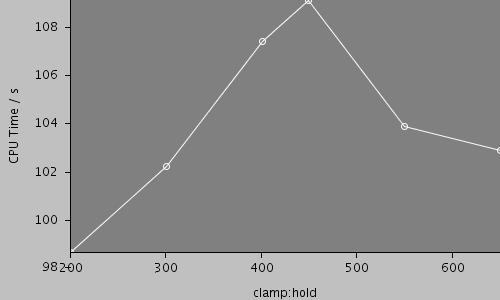
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | Non Gated channels / cpmts | time/ms | timestep/ms | clamp:hold | CPU Time / s |
| 517 | 624301 / 516 | 349976 / 19 | 0 / 0 | 2.00e+03 | 0.05 | 200 | 99.3 |
| 517 | 624301 / 516 | 349976 / 19 | 0 / 0 | 2.00e+03 | 0.05 | 300 | 103 |
| 517 | 624301 / 516 | 349976 / 19 | 0 / 0 | 2.00e+03 | 0.05 | 400 | 107 |
| 517 | 624301 / 516 | 349976 / 19 | 0 / 0 | 2.00e+03 | 0.05 | 450 | 109 |
| 517 | 624301 / 516 | 349976 / 19 | 0 / 0 | 2.00e+03 | 0.05 | 550 | 107 |
| 517 | 624301 / 516 | 349976 / 19 | 0 / 0 | 2.00e+03 | 0.05 | 650 | 103 |
 |
Morphology: null






Predefined views
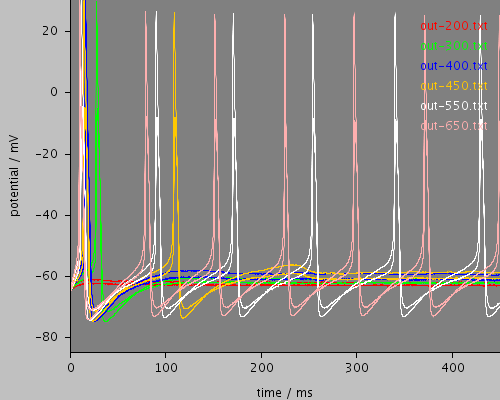
whole
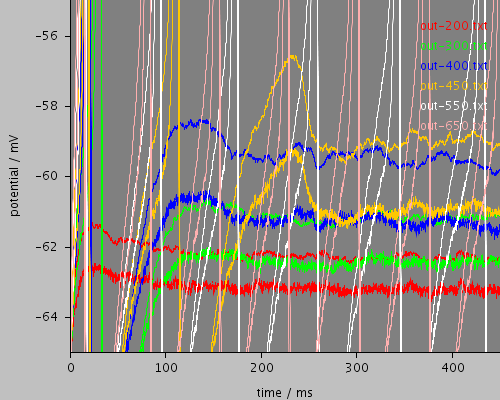
wholethreshold
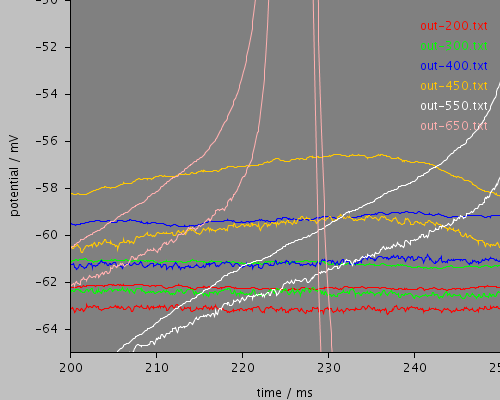
enlarged
650
All files
Model
Archive file of the complete model: mainen.jarrun.xml
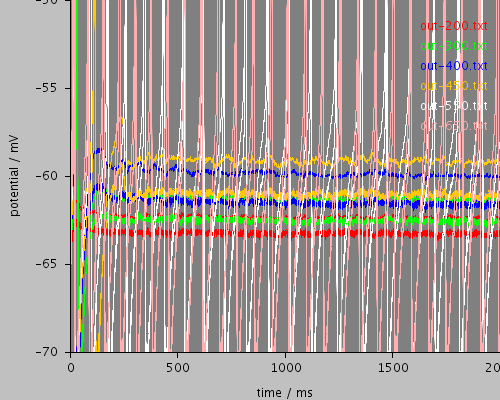
<PSICSRun timeStep="0.05ms" runTime="2000ms" startPotential="-65mV" environment="environment" properties="membrane" access="recording" stochThreshold="10000"> <MorphologySource format="swc" file="NM1.swc"/> <StructureDiscretization baseElementSize="15um"/> <RunSet vary="clamp:hold" values="[200, 300, 400, 450, 550, 650]pA" filepattern="out-$"/> <ViewConfig> <LineGraph width="500" height="400"> <XAxis min="0" max="250" label="time / ms"/> <YAxis min="-80" max="60" label="potential / mV"/> <LineSet file="out-200.txt" color="red"/> <LineSet file="out-300.txt" color="green"/> <LineSet file="out-400.txt" color="blue"/> <LineSet file="out-450.txt" color="orange"/> <LineSet file="out-550.txt" color="white"/> <LineSet file="out-650.txt" color="pink"/> <View id="whole" xmin="-10." xmax="450." ymin="-85." ymax="30."/> <View id="wholethreshold" xmin="-10." xmax="450." ymin="-65." ymax="-55."/> <View id="enlarged" xmin="200." xmax="250." ymin="-65." ymax="-50."/> <View id="650" xmin="0." xmax="2000." ymin="-70." ymax="-50."/> </LineGraph> </ViewConfig> </PSICSRun>
membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="100ohm_cm"
membraneCapacitance="1uF_per_cm2">
<ChannelPopulation channel="leak-Na" density="0.1per_um2"/>
<ChannelPopulation channel="leak-K" density="0.1per_um2"/>
<ChannelPopulation channel="naz_nature" density="60per_um2" distribution="proximal"/>
<ChannelPopulation channel="kvz_nature" density="18per_um2" distribution="proximal"/>
<ChannelPopulation channel="km" density="0.1per_um2" distribution="proximal"/>
<DistributionRule id="proximal">
<RegionMask action="include" where="p .lt. 100"/>
</DistributionRule>
<DensityAdjustment maintain="-65mV" vary="leak-Na, leak-K"/>
</CellProperties>
environment.xml
<CellEnvironment id="environment" temperature="13Celsius"> <Ion id="Na" name="Sodium" reversalPotential="40mV"/> <Ion id="K" name="Potassium" reversalPotential="-80mV"/> </CellEnvironment>
recording.xml
<Access id="recording"> <CellLocation id="p0" path="10" rankBy="radius" sequenceFraction="1."/> <CellLocation id="p1" path="120" rankBy="radius" sequenceFraction="0.5"/> <CellLocation id="p2" path="240" rankBy="radius" sequenceFraction="0.5"/> <CurrentClamp id="clamp" location="p0" lineColor="red" hold="0pA"/> <VoltageRecorder location="p2" lineColor="blue"/> </Access>
leak-Na.xml
<KSChannel id="leak-Na" gSingle="20pS" permeantIon="Na"> <OpenState id="o1"/> <ClosedState id="c1"/> <FixedRateTransition from="o1" to="c1" forward="3.per_ms" reverse="7.per_ms"/> </KSChannel>
leak-K.xml
<KSChannel id="leak-K" gSingle="20pS" permeantIon="K"> <OpenState id="o1"/> <ClosedState id="c1"/> <FixedRateTransition from="o1" to="c1" forward="3.per_ms" reverse="7.per_ms"/> </KSChannel>
naz_nature.xml
<KSChannel id="naz_nature" permeantIon="Na" gSingle="20pS">
<About>
Converted from naz_nature.mod (Zack Mainen, 1994)
</About>
<KSComplex id="m" instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<ExpLinearTransition from="c" to="o" rate="1.64per_ms" midpoint ="-35.mV" scale="9mV"
baseTemperature="23Celsius" q10="2.3"/>
<ExpLinearTransition from="o" to="c" rate="1.12per_ms" midpoint="-35.mV" scale="-9mV"
baseTemperature="23Celsius" q10="2.3"/>
</KSComplex>
<CodedTransitionFunction name="trap0" returnVariable="rate" type="double">
<Argument name="v" type="double"/>
<Argument name="th" type="double"/>
<Argument name="a" type="double"/>
<Argument name="q" type="double"/>
<![CDATA[
if (Math.abs(v - th) > 1.e-6) {
rate = a * (v - th) / (1 - Math.exp(-(v - th)/q));
} else {
rate = a * q;
}
]]>
</CodedTransitionFunction>
<KSComplex id="h">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="htau" infvar="hinf"
baseTemperature="23Celsius" q10="2.3">
<Constant name="thi1" value="-50"/>
<Constant name="thi2" value="-75"/>
<Constant name ="rd" value="0.024"/>
<Constant name="rg" value="0.0091"/>
<Constant name="qi" value="5"/>
<Constant name="thinf" value="-65"/>
<Constant name="qinf" value="6.2"/>
<![CDATA[
double a = trap0(v, thi1, rd, qi);
double b = trap0(-v, -thi2, rg, qi);
htau = 1/(a+b);
hinf = 1. / (1 + Math.exp((v - thinf)/qinf));
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
kvz_nature.xml
<KSChannel id="kvz_nature" permeantIon="K" gSingle="20pS"> <About> Converted from kvz_nature.mod (Zach Mainen, 1995) </About> <KSComplex id="n" instances="1"> <ClosedState id="c"/> <OpenState id="o"/> <ExpLinearTransition from="c" to="o" rate="0.18per_ms" midpoint ="25.mV" scale="9mV" baseTemperature="23Celsius" q10="2.3"/> <ExpLinearTransition from="o" to="c" rate="0.018per_ms" midpoint ="25.mV" scale="-9mV" baseTemperature="23Celsius" q10="2.3"/> </KSComplex> </KSChannel>
km.xml
<KSChannel id="km" permeantIon="K" gSingle="20pS"> <About> Converted from km.mod, slow non-inactivating potassium current based on I-M (Zach Mainen, 1995) </About> <KSComplex id="n" instances="1"> <ClosedState id="c"/> <OpenState id="o"/> <ExpLinearTransition from="c" to="o" rate="0.009per_ms" midpoint ="-30.mV" scale="9mV" baseTemperature="23Celsius" q10="2.3"/> <ExpLinearTransition from="o" to="c" rate="0.0009per_ms" midpoint ="-30.mV" scale="-9mV" baseTemperature="23Celsius" q10="2.3"/> </KSComplex> </KSChannel>