stimtest (run.xml)
Current clamp stimulation profile read from a file, varying the stimulation scale factor
Total CPU time 0.1200 seconds; at 11:55:00 Thu 24 Sep 2009
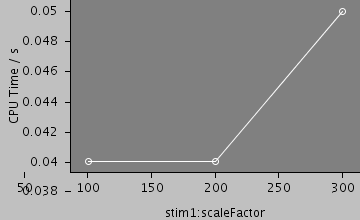
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | Non Gated channels / cpmts | time/ms | timestep/ms | stim1:scaleFactor | CPU Time / s |
| 159 | 0 / 0 | 0 / 0 | 0 / 0 | 25.0 | 0.01 | 100 | 0.0400 |
| 159 | 0 / 0 | 0 / 0 | 0 / 0 | 25.0 | 0.01 | 200 | 0.0400 |
| 159 | 0 / 0 | 0 / 0 | 0 / 0 | 25.0 | 0.01 | 300 | 0.0400 |
 |
Morphology: null
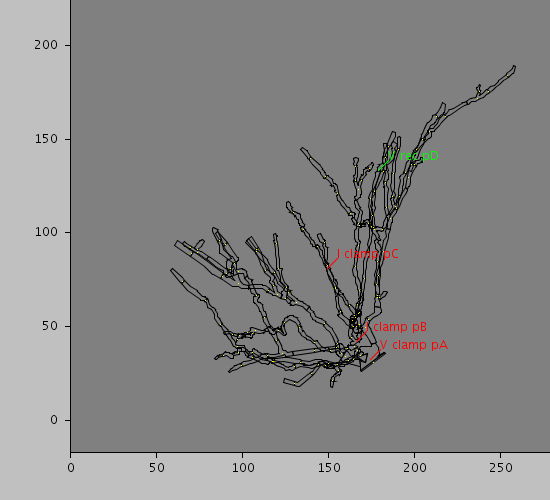
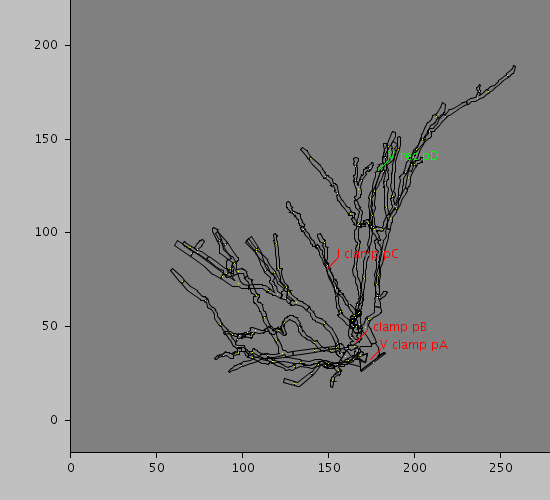
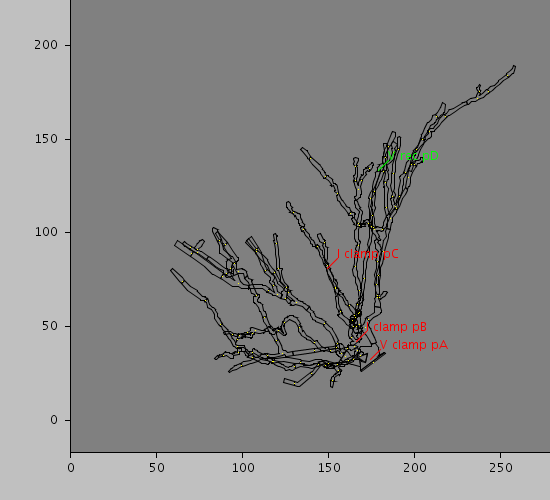
Predefined views
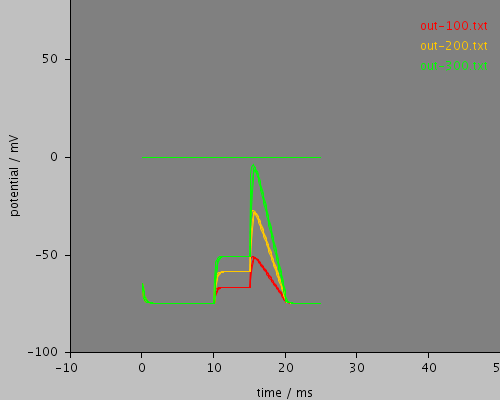
whole
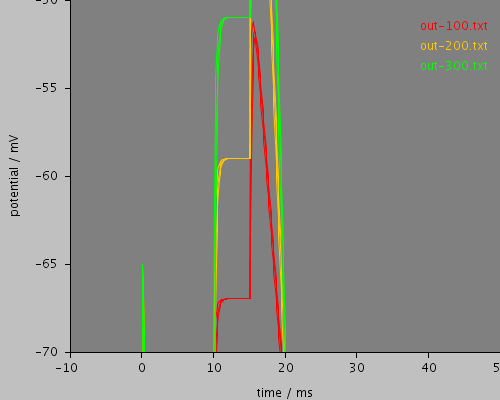
enlarged
All files
| Model | Preprocessed | Outupt data | Reference data etc |
| run.xml membrane.xml environment.xml recording.xml |
out-100.ppp out-200.ppp out-300.ppp |
log.txt out-100.sum out-200.sum out-300.sum out-100.dat out-100.txt out-200.dat out-200.txt out-300.dat out-300.txt |
stim1.txt NM1.swc out-merged.txt |
Model
Archive file of the complete model: stimtest.jarrun.xml
<PSICSRun timeStep="0.01ms" runTime="25ms" startPotential="-65mV" environment="environment" properties="membrane" access="recording" stochThreshold="0"> <MorphologySource format="swc" file="NM1b.swc"/> <StructureDiscretization baseElementSize="12um"/> <RunSet vary="stim1:scaleFactor" values="[100, 200, 300]" filepattern="out-$" merge="true"/> <!-- nb this doesn't work - can't parse string arrays for values <RunSet vary="stim1:file" values="[stim1.txt, stim2.txt]" filepattern="out-$" merge="true"/> --> <info>Current clamp stimulation profile read from a file, varying the stimulation scale factor</info> <ViewConfig> <LineGraph width="500" height="400"> <XAxis min="0" max="250" label="time / ms"/> <YAxis min="-80" max="60" label="potential / mV"/> <LineSet file="out-100.txt" color="red"/> <LineSet file="out-200.txt" color="orange"/> <LineSet file="out-300.txt" color="green"/> <View id="whole" xmin="-10." xmax="50." ymin="-100." ymax="80."/> <View id="enlarged" xmin="-10." xmax="50." ymin="-70." ymax="-50."/> </LineGraph> </ViewConfig> </PSICSRun>
membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="100ohm_cm"
membraneCapacitance="1uF_per_cm2">
</CellProperties>
environment.xml
<CellEnvironment id="environment"> <Ion id="Na" name="Sodium" reversalPotential="40mV"/> <Ion id="K" name="Potassium" reversalPotential="-80mV"/> </CellEnvironment>
recording.xml
<Access id="recording" recordClamps="true">
<CellLocation id="pA" path="10" rankBy="radius" sequenceFraction="1."/>
<CellLocation id="pB" path="50" rankBy="radius" sequenceFraction="1."/>
<CellLocation id="pC" path="120" rankBy="radius" sequenceIndex="0"/>
<CellLocation id="pD" path="160" rankBy="radius" sequenceFraction="0.5"/>
<VoltageClamp location="pA" hold="-75mV"/>
<CurrentClamp location="pB" lineColor="red" hold="0.0nA">
<TimeSeries id="stim1" file="stim1.txt" timeUnit="1ms" valueUnit="0.01nA"
scaleFactor="1.0"/>
</CurrentClamp>
<CurrentClamp location="pC" lineColor="red" hold="0.0nA">
<TimeSeries id="stim2" file="stim1.txt" column="2" timeUnit="1ms" valueUnit="0.01nA"
scaleFactor="100.0"/>
</CurrentClamp>
<VoltageRecorder location="pD" lineColor="blue"/>
</Access>