migliore-1c (run1c.xml)
Migliore et al CA1 pyramidal cell
Total CPU time 2.551 seconds; at 15:28:43 Wed 19 Sep 2007
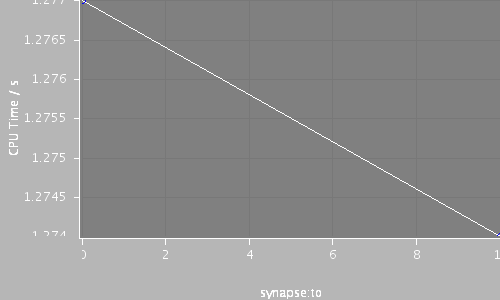
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | time/ms | timestep/ms | synapse:to | CPU Time / s |
| 320 | 0 / 0 | 677777 / 224 | 120.0 | 0.025 | 0 | 1.28 |
| 320 | 0 / 0 | 677775 / 222 | 120.0 | 0.025 | 10 | 1.274 |
 |
Predefined views
whole
first
later
All files
Model
Archive file of the complete model: migliore-1c.jarrun1c.xml
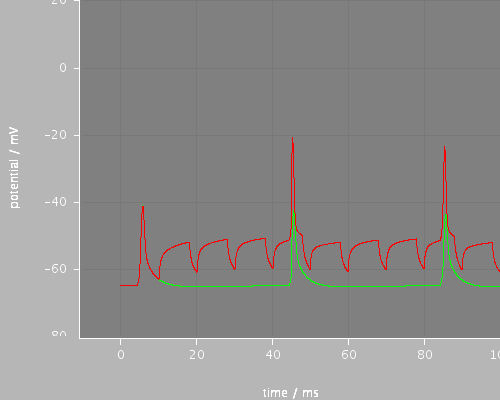
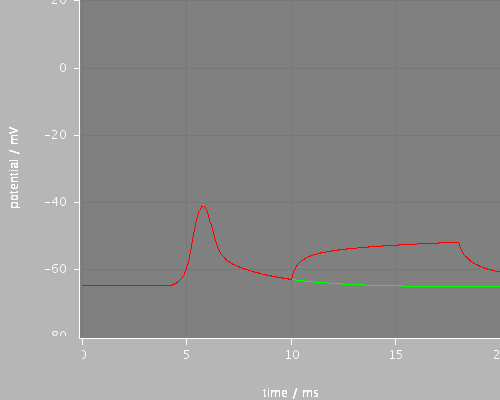
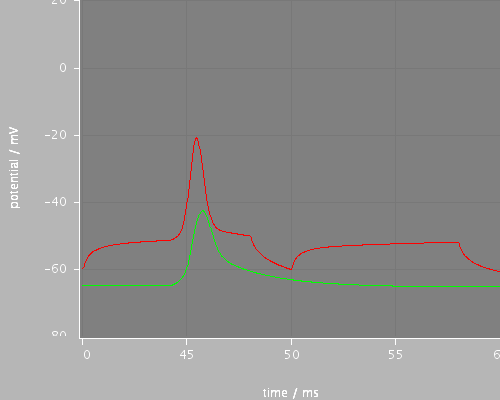
<PSICSRun timeStep="0.025ms" runTime="120ms" startPotential="-65mV" morphology="morph59" environment="environment" properties="membrane" access="recording-c" stochThreshold="0"> <StructureDiscretization baseElementSize="10um"/> <info>Migliore et al CA1 pyramidal cell</info> <RunSet vary="synapse:to" values="[0, 10]nS" filepattern="out-$.txt"/> <ViewConfig> <LineGraph width="500" height="400"> <XAxis min="0" max="250" label="time / ms"/> <YAxis min="-80" max="60" label="potential / mV"/> <LineSet file="out-0-f.txt" color="green" show="2"/> <LineSet file="out-10-f.txt" color="red" show="2"/> <View id="whole" xmin="-10." xmax="100." ymin="-80." ymax="20."/> <View id="first" xmin="0" xmax="20." ymin="-80." ymax="20."/> <View id="later" xmin="40" xmax="60." ymin="-80." ymax="20."/> </LineGraph> </ViewConfig> </PSICSRun>
morph59.xml (truncated)
<morphml id="morph59">
<!-- XML file generated by NEURON 5.9 ModelViewer -->
<!-- Authors: Michael Hines and Sushil Kambampati -->
<!-- Yale University -->
<!-- Date: Fri Sep 7 12:52:05 BST 2007 -->
<cells>
<cell name="soma[0]">
<segments>
<segment id="1" name = "Seg0_soma[0]" cable = "0">
<proximal x="0" y="0" z="0" diameter="3.4"/>
<distal x="0.25" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="2" name = "Seg1_soma[0]" parent="1" cable = "0">
<distal x="0.5" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="671" name = "Seg0_basal[41]" parent="1" cable = "181">
<proximal x="0" y="0" z="0" diameter="1"/>
<distal x="-4.38737" y="1.85495" z="0" diameter="1"/>
</segment>
<segment id="672" name = "Seg1_basal[41]" parent="671" cable = "181">
<distal x="-8.77474" y="3.7099" z="0" diameter="1"/>
</segment>
<segment id="606" name = "Seg0_basal[23]" parent="1" cable = "163">
<proximal x="0" y="0" z="0" diameter="1"/>
<distal x="-5.95875" y="0.799243" z="0" diameter="1"/>
</segment>
<segment id="607" name = "Seg1_basal[23]" parent="606" cable = "163">
<distal x="-11.9175" y="1.59849" z="0" diameter="1"/>
</segment>
<segment id="593" name = "Seg0_basal[20]" parent="1" cable = "160">
<proximal x="0" y="0" z="0" diameter="0.4"/>
<distal x="-8.98517" y="-1.20517" z="0" diameter="0.4"/>
</segment>
<segment id="594" name = "Seg1_basal[20]" parent="593" cable = "160">
<distal x="-26.9555" y="-3.61552" z="0" diameter="0.4"/>
</segment>
<segment id="595" name = "Seg2_basal[20]" parent="594" cable = "160">
<distal x="-35.9407" y="-4.82069" z="0" diameter="0.4"/>
</segment>
<segment id="517" name = "Seg0_basal[0]" parent="1" cable = "140">
<proximal x="0" y="0" z="0" diameter="0.8"/>
<distal x="-1.33023" y="-0.562414" z="0" diameter="0.8"/>
</segment>
<segment id="518" name = "Seg1_basal[0]" parent="517" cable = "140">
<distal x="-2.66047" y="-1.12483" z="0" diameter="0.8"/>
</segment>
<segment id="3" name = "Seg0_soma[1]" parent="2" cable = "1">
<proximal x="0.5" y="0" z="0" diameter="3.4"/>
<distal x="0.55" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="4" name = "Seg1_soma[1]" parent="3" cable = "1">
<distal x="0.6" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="736" name = "Seg0_basal[59]" parent="672" cable = "199">
<proximal x="-8.77474" y="3.7099" z="0" diameter="0.4"/>
<distal x="-15.8252" y="10.9693" z="0" diameter="0.4"/>
</segment>
<segment id="737" name = "Seg1_basal[59]" parent="736" cable = "199">
<distal x="-29.9261" y="25.4881" z="0" diameter="0.4"/>
</segment>
- and 2829 more lines -membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="150ohm_cm"
membraneCapacitance="1.0uF_per_cm2">
<Exclusion either="*apical*" or="*soma*"/>
<PassiveProperties region="*apical*" membraneCapacitance="2uF_per_cm2"/>
<PassiveProperties region="*basal*" membraneCapacitance="2uF_per_cm2"/>
<PassiveProperties region="*axon*" cytoplasmResistivity="50ohm_cm"/>
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<CellRegion match="*axon*">
<ChannelPopulation channel="Nax-coded" density="21.33per_um2"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="3.2per_um2"/>
</CellRegion>
<CellRegion match="*soma*">
<ChannelPopulation channel="Na3-coded-soma" density="10.67per_um2"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2"/>
</CellRegion>
<CellRegion match="*basal*">
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<ChannelPopulation channel="Na3-coded-basal" density="10.67per_um2"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2"/>
</CellRegion>
<CellRegion match="*apical*">
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<ChannelPopulation channel="Na3-coded-apical" density="10.67per_um2" distribution="fat500"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2" distribution="fat500"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2" distribution="kaprox"/>
<ChannelPopulation channel="Kadist-coded" density="16per_um2" distribution="kadist"/>
</CellRegion>
<DistributionRule id="fat500">
<RegionMask action="exclude" where="diameter .lt. 0.5"/>
<RegionMask action="exclude" where="path .gt. 500"/>
</DistributionRule>
<DistributionRule id="kaprox" expression="(1 + path / 100)">
<RegionMask action="exclude" where="path .gt. 100"/>
</DistributionRule>
<DistributionRule id="kadist" expression="(1 + path / 100)">
<RegionMask action="exclude" where="path .lt. 100"/>
<RegionMask action="exclude" where="path .gt. 500"/>
</DistributionRule>
<DensityAdjustment maintain="-65mV" vary="leak-Na, leak-K"/>
</CellProperties>
environment.xml
<CellEnvironment id="environment" temperature="34Celsius"> <Ion id="K" name="Potassium" reversalPotential="-90mV"/> <Ion id="Na" name="Sodium" reversalPotential="55mV"/> </CellEnvironment>
recording-c.xml
<Access id="recording-c"> <CurrentClamp at="4" hold="0nA"> <CurrentPulse start="4ms" duration="1.2ms" to="2nA"/> <CurrentPulse start="44ms" duration="1.2ms" to="2nA"/> <CurrentPulse start="84ms" duration="1.2ms" to="2nA"/> </CurrentClamp> <ConductanceClamp id="ccc" at="Seg0_apical[13]" potential="0mV"> <ConductancePulse id="synapse" start="10ms" duration="8ms" to="0nS" repeatAfter="10ms"/> </ConductanceClamp> <VoltageRecorder at="Seg0_apical[12]"/> </Access>
Nax-coded.xml
<KSChannel id="Nax-coded" permeantIon="Na" gSingle="30pS">
<CodedTransitionFunction name="trap0" returnVariable="rate" type="double">
<Argument name="v" type="double"/>
<Argument name="th" type="double"/>
<Argument name="a" type="double"/>
<Argument name="q" type="double"/>
<![CDATA[
if (Math.abs(v - th) > 1.e-6) {
rate = a * (v - th) / (1 - Math.exp(-(v - th)/q));
} else {
rate = a * q;
}
]]>
</CodedTransitionFunction>
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="mtau" infvar="minf">
<Constant name="q10" value="2"/>
<Constant name="tha" value="-30" info="vhalf, mV"/>
<Constant name="Ra" value="0.4" info="opening rate, per ms"/>
<Constant name="qa" value="7.2" info="activation slope, mV"/>
<Constant name="Rb" value="0.124" info="closing rate, per ms"/>
<Constant name="mmin" value="0.02" info="minimum time constant"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v ,tha,Ra,qa);
double b = trap0(-v ,-tha,Rb,qa);
mtau = 1 / (a + b) / qt;
if (mtau < mmin) mtau=mmin;
minf = a / (a + b);
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="htau" infvar="hinf">
<Constant name="q10" value="2"/>
<Constant name="thi1" value="-45" info="vhalf, mV"/>
<Constant name="thi2" value="-45" info="vhalf, mV"/>
<Constant name="Rd" value="0.03" info="opening rate, per ms"/>
<Constant name="Rg" value="0.01" info="closing rate, per ms"/>
<Constant name="qd" value="1.5" info="activation slope, mV"/>
<Constant name="qg" value="1.5" info="activation slope, mV"/>
<Constant name="hmin" value="0.5" info="minimum time constant"/>
<Constant name="thinf" value="-50" info="inact inf slope, mV"/>
<Constant name="qinf" value="4"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v, thi1,Rd,qd);
double b = trap0(-v, -thi2,Rg,qg);
htau = 1 / (a + b) / qt;
if (htau < hmin) htau=hmin;
hinf = 1/(1+Math.exp((v-thinf)/qinf));
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Kdr-coded.xml
<KSChannel id="Kdr-coded" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="ntau" infvar="ninf">
<Constant name="q10" value="1"/>
<Constant name="zetan" value="-3"/>
<Constant name="vhalfn" value="13" info="mV"/>
<Constant name="a0n" value="0.02"/>
<Constant name="gmn" value="0.7"/>
<Constant name="nmin" value="2"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double a = Math.exp(1.e-3*zetan*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ninf = 1/(1 + a);
double betn = Math.exp(1.e-3*zetan*gmn*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ntau = betn /(qt*a0n*(1+a));
if (ntau < nmin) ntau = nmin;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Kaprox-coded.xml
<KSChannel id="Kaprox-coded" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="ntau" infvar="ninf">
<Constant name="q10" value="5"/>
<Constant name="zetan" value="-1.5"/>
<Constant name="pw" value="-1" info=""/>
<Constant name="tq" value="-40" info=""/>
<Constant name="qq" value="5" info=""/>
<Constant name="vhalfn" value="11" info="mV"/>
<Constant name="gmn" value="0.55"/>
<Constant name="nmin" value="0.1" info="minimum time constant"/>
<Constant name="a0n" value="0.05"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double zeta=zetan+pw/(1+Math.exp((v-tq)/qq));
double a = Math.exp(1.e-3*zeta*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ninf = 1/(1 + a);
double betn = Math.exp(1.e-3*zeta*gmn*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ntau = betn /(qt*a0n*(1+a));
if (ntau < nmin) ntau = nmin;
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<OpenState id="o"/>
<ClosedState id="c"/>
<TauInfCodedTransition from="c" to="o" tauvar="ltau" infvar="linf">
<Constant name="q10" value="5"/>
<Constant name="zetal" value="3"/>
<Constant name="vhalfl" value="-56" info="mV"/>
<Constant name="qtl" value="1" info=""/>
<Constant name="lmin" value="2" info="minimum time constant"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double a = Math.exp(1.e-3*zetal*(v-vhalfl)*9.648e4/(8.315*(273.16+temperature)));
linf = 1 / (1 + a);
ltau = 0.26 * (v + 50) / qtl;
if (ltau < lmin / qtl) ltau = lmin / qtl;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Na3-coded-soma.xml
<KSChannel id="Na3-coded-soma" permeantIon="Na" gSingle="30pS">
<CodedTransitionFunction name="trap0" returnVariable="rate" type="double">
<Argument name="v" type="double"/>
<Argument name="th" type="double"/>
<Argument name="a" type="double"/>
<Argument name="q" type="double"/>
<![CDATA[
if (Math.abs(v - th) > 1.e-6) {
rate = a * (v - th) / (1 - Math.exp(-(v - th)/q));
} else {
rate = a * q;
}
]]>
</CodedTransitionFunction>
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="mtau" infvar="minf">
<Constant name="q10" value="2"/>
<Constant name="tha" value="-30"/>
<Constant name ="Ra" value="0.4"/>
<Constant name="Rb" value="0.124"/>
<Constant name="qa" value="7.2"/>
<Constant name="mmin" value="0.02"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v, tha,Ra,qa);
double b = trap0(-v, -tha,Rb,qa);
mtau = 1/(a+b)/qt;
if (mtau<mmin) mtau=mmin;
minf = a/(a+b);
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="htau" infvar="hinf">
<Constant name="q10" value="2"/>
<Constant name="thi1" value="-45"/>
<Constant name="thi2" value="-45"/>
<Constant name="thinf" value="-50"/>
<Constant name ="Rd" value="0.03"/>
<Constant name="Rg" value="0.01"/>
<Constant name="qd" value="1.5"/>
<Constant name="qg" value="1.5"/>
<Constant name="hmin" value="0.5"/>
<Constant name="qinf" value="4"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v,thi1,Rd,qd);
double b = trap0(-v,-thi2,Rg,qg);
htau = 1/(a+b)/qt;
if (htau<hmin) htau=hmin;
hinf = 1/(1+Math.exp((v-thinf)/qinf));
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="stau" infvar="sinf">
<Constant name="q10" value="2"/>
<Constant name="vvh" value="-58"/>
<Constant name="vvs" value="2"/>
<Constant name ="a2" value="0.8" id="Na3-a2-semiconstant"/> <!-- a2 is the only change between soma, apical basal. -->
<Constant name="zetas" value="12"/>
<Constant name="gms" value="0.2"/>
<Constant name="vhalfs" value="-60"/>
<Constant name="a0s" value="0.0003"/>
<Constant name="smax" value="10"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double c = 1 / (1+Math.exp((v-vvh)/vvs));
sinf = c+a2*(1-c);
double bets = Math.exp(1.e-3*zetas*gms*(v-vhalfs)*9.648e4/(8.315*(273.16+temperature)));
double alps = Math.exp(1.e-3*zetas*(v-vhalfs)*9.648e4/(8.315*(273.16+temperature)));
stau = bets / (a0s * (1 + alps));
if (stau<smax) stau=smax;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
leak-Na.xml
<KSChannel id="leak-Na" gSingle="1pS" permeantIon="Na"> <OpenState id="o1"/> </KSChannel>
leak-K.xml
<KSChannel id="leak-K" gSingle="1pS" permeantIon="K"> <OpenState id="o1"/> </KSChannel>
Na3-coded-basal.xml
<DerivedKSChannel id="Na3-coded-basal" from="Na3-coded-soma"> <ParameterChange to="Na3-a2-semiconstant" attribute="value" newText="1"/> </DerivedKSChannel>
Na3-coded-apical.xml
<DerivedKSChannel id="Na3-coded-apical" from="Na3-coded-soma"> <ParameterChange to="Na3-a2-semiconstant" attribute="value" newText="0.5"/> </DerivedKSChannel>
Kadist-coded.xml
<KSChannel id="Kadist-coded" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="ntau" infvar="ninf">
<Constant name="q10" value="5"/>
<Constant name="zetan" value="-1.8"/>
<Constant name="pw" value="-1" info=""/>
<Constant name="tq" value="-40" info=""/>
<Constant name="qq" value="5" info=""/>
<Constant name="vhalfn" value="-1" info="mV"/>
<Constant name="gmn" value="0.39"/>
<Constant name="nmin" value="0.2" info="minimum time constant"/>
<Constant name="a0n" value="0.1"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double zeta=zetan+pw/(1+Math.exp((v-tq)/qq));
double a = Math.exp(1.e-3*zeta*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ninf = 1/(1 + a);
double betn = Math.exp(1.e-3*zeta*gmn*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ntau = betn /(qt*a0n*(1+a));
if (ntau < nmin) ntau = nmin;
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<OpenState id="o"/>
<ClosedState id="c"/>
<TauInfCodedTransition from="c" to="o" tauvar="ltau" infvar="linf">
<Constant name="q10" value="5"/>
<Constant name="zetal" value="3"/>
<Constant name="vhalfl" value="-56" info="mV"/>
<Constant name="qtl" value="1" info=""/>
<Constant name="lmin" value="2" info="minimum time constant"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double a = Math.exp(1.e-3*zetal*(v-vhalfl)*9.648e4/(8.315*(273.16+temperature)));
linf = 1 / (1 + a);
ltau = 0.26 * (v + 50) / qtl;
if (ltau < lmin / qtl) ltau = lmin / qtl;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>