migliore-devel (run1c.xml)
Migliore et al CA1 pyramidal cell
Total CPU time 2.609 seconds; at 16:24:58 Tue 18 Sep 2007
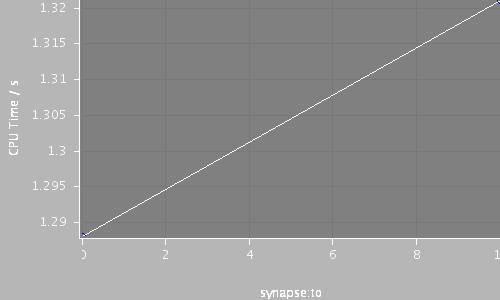
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | time/ms | timestep/ms | synapse:to | CPU Time / s |
| 320 | 0 / 0 | 683051 / 223 | 120.0 | 0.025 | 0 | 1.288 |
| 320 | 0 / 0 | 683053 / 224 | 120.0 | 0.025 | 10 | 1.321 |
 |
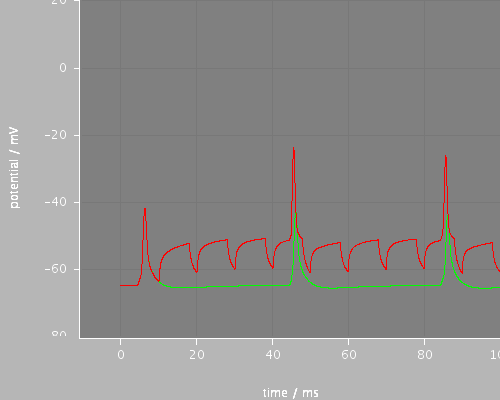
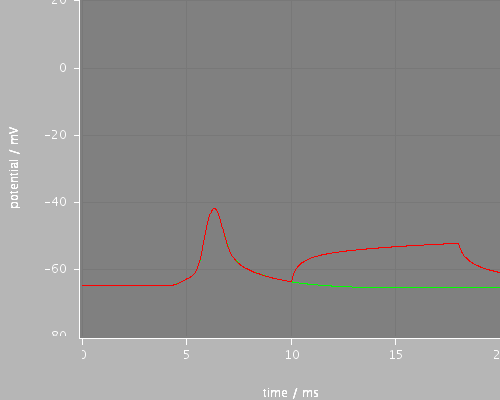
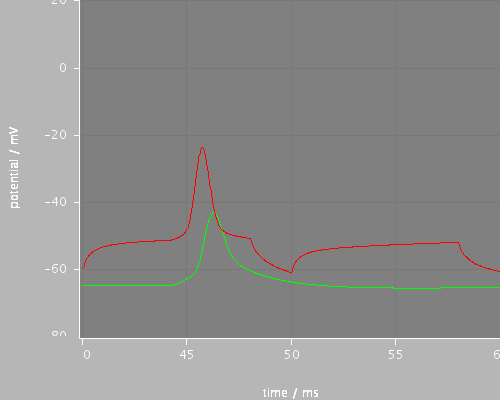
Predefined views
whole
first
later
All files
Model
Archive file of the complete model: migliore-devel.jarcodetestrun.xml
<PSICSRun timeStep="0.1ms" runTime="120ms" startPotential="-65mV" morphology="morph59" environment="environment" properties="codetestprops" access="codetestrec" stochThreshold="0"> <StructureDiscretization baseElementSize="10um"/> <info>testing the coded channels</info> <ViewConfig> <LineGraph width="500" height="400"> <XAxis min="0" max="250" label="time / ms"/> <YAxis min="-80" max="60" label="potential / mV"/> <LineSet file="psics-out-f.txt" color="green"/> <View id="whole" xmin="-10." xmax="120." ymin="-80." ymax="20."/> <View id="first" xmin="5" xmax="20." ymin="-80." ymax="20."/> </LineGraph> </ViewConfig> </PSICSRun>
cablecell.xml
<CellMorphology id="cablecell"> <Point id="a" x="0" y="0" z="0" r="0.5"/> <Point parent="a" id="b" x="1000" y="0" z = "0" r="0.5"/> </CellMorphology>
codetestprops.xml
<CellProperties id="codetestprops"
cytoplasmResistivity="100ohm_cm"
membraneCapacitance="1uF_per_cm2">
<PassiveProperties region="apical" membraneCapacitance="2uF_per_cm2" cytoplasmResistivity="200ohm_cm"/>
<ChannelPopulation channel="leak-Na" density="1per_um2"/>
<ChannelPopulation channel="leak-K" density="1per_um2"/>
<ChannelPopulation channel="Nax-coded" density="12per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="25per_um2"/>
<ChannelPopulation channel="Na3-coded-soma" density="1per_um2"/>
<ChannelPopulation channel="Na3-coded-apical" density="1per_um2"/>
<ChannelPopulation channel="Na3-coded-basal" density="1per_um2"/>
<ChannelPopulation channel="Naxfit-coded" density="1per_um2"/>
<ChannelPopulation channel="Naxfit" density="1per_um2"/>
<DensityAdjustment maintain="-65mV" vary="leak-Na, leak-K"/>
</CellProperties>
environment.xml
<CellEnvironment id="environment" temperature="34Celsius"> <Ion id="K" name="Potassium" reversalPotential="-90mV"/> <Ion id="Na" name="Sodium" reversalPotential="55mV"/> </CellEnvironment>
codetestrec.xml
<Access id="codetestrec"> <CurrentClamp at="1" hold="0.15nA"> </CurrentClamp> <VoltageRecorder at="100"/> </Access>
Nax-coded.xml
<KSChannel id="Nax-coded" permeantIon="Na" gSingle="30pS">
<CodedTransitionFunction name="trap0" returnVariable="rate" type="double">
<Argument name="v" type="double"/>
<Argument name="th" type="double"/>
<Argument name="a" type="double"/>
<Argument name="q" type="double"/>
<![CDATA[
if (Math.abs(v - th) > 1.e-6) {
rate = a * (v - th) / (1 - Math.exp(-(v - th)/q));
} else {
rate = a * q;
}
]]>
</CodedTransitionFunction>
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="mtau" infvar="minf">
<Constant name="q10" value="2"/>
<Constant name="tha" value="-30" info="vhalf, mV"/>
<Constant name="Ra" value="0.4" info="opening rate, per ms"/>
<Constant name="qa" value="7.2" info="activation slope, mV"/>
<Constant name="Rb" value="0.124" info="closing rate, per ms"/>
<Constant name="mmin" value="0.02" info="minimum time constant"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v ,tha,Ra,qa);
double b = trap0(-v ,-tha,Rb,qa);
mtau = 1 / (a + b) / qt;
if (mtau < mmin) mtau=mmin;
minf = a / (a + b);
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="htau" infvar="hinf">
<Constant name="q10" value="2"/>
<Constant name="thi1" value="-45" info="vhalf, mV"/>
<Constant name="thi2" value="-45" info="vhalf, mV"/>
<Constant name="Rd" value="0.03" info="opening rate, per ms"/>
<Constant name="Rg" value="0.01" info="closing rate, per ms"/>
<Constant name="qd" value="1.5" info="activation slope, mV"/>
<Constant name="qg" value="1.5" info="activation slope, mV"/>
<Constant name="hmin" value="0.5" info="minimum time constant"/>
<Constant name="thinf" value="-50" info="inact inf slope, mV"/>
<Constant name="qinf" value="4"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v, thi1,Rd,qd);
double b = trap0(-v, -thi2,Rg,qg);
htau = 1 / (a + b) / qt;
if (htau < hmin) htau=hmin;
hinf = 1/(1+Math.exp((v-thinf)/qinf));
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Kaprox-coded.xml
<KSChannel id="Kaprox-coded" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="ntau" infvar="ninf">
<Constant name="q10" value="5"/>
<Constant name="zetan" value="-1.5"/>
<Constant name="pw" value="-1" info=""/>
<Constant name="tq" value="-40" info=""/>
<Constant name="qq" value="5" info=""/>
<Constant name="vhalfn" value="11" info="mV"/>
<Constant name="gmn" value="0.55"/>
<Constant name="nmin" value="0.1" info="minimum time constant"/>
<Constant name="a0n" value="0.05"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double zeta=zetan+pw/(1+Math.exp((v-tq)/qq));
double a = Math.exp(1.e-3*zeta*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ninf = 1/(1 + a);
double betn = Math.exp(1.e-3*zeta*gmn*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ntau = betn /(qt*a0n*(1+a));
if (ntau < nmin) ntau = nmin;
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<OpenState id="o"/>
<ClosedState id="c"/>
<TauInfCodedTransition from="c" to="o" tauvar="ltau" infvar="linf">
<Constant name="q10" value="5"/>
<Constant name="zetal" value="3"/>
<Constant name="vhalfl" value="-56" info="mV"/>
<Constant name="qtl" value="1" info=""/>
<Constant name="lmin" value="2" info="minimum time constant"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double a = Math.exp(1.e-3*zetal*(v-vhalfl)*9.648e4/(8.315*(273.16+temperature)));
linf = 1 / (1 + a);
ltau = 0.26 * (v + 50) / qtl;
if (ltau < lmin / qtl) ltau = lmin / qtl;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
leak-Na.xml
<KSChannel id="leak-Na" gSingle="1pS" permeantIon="Na"> <OpenState id="o1"/> </KSChannel>
leak-K.xml
<KSChannel id="leak-K" gSingle="1pS" permeantIon="K"> <OpenState id="o1"/> </KSChannel>
Na3-coded-soma.xml
<KSChannel id="Na3-coded-soma" permeantIon="Na" gSingle="30pS">
<CodedTransitionFunction name="trap0" returnVariable="rate" type="double">
<Argument name="v" type="double"/>
<Argument name="th" type="double"/>
<Argument name="a" type="double"/>
<Argument name="q" type="double"/>
<![CDATA[
if (Math.abs(v - th) > 1.e-6) {
rate = a * (v - th) / (1 - Math.exp(-(v - th)/q));
} else {
rate = a * q;
}
]]>
</CodedTransitionFunction>
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="mtau" infvar="minf">
<Constant name="q10" value="2"/>
<Constant name="tha" value="-30"/>
<Constant name ="Ra" value="0.4"/>
<Constant name="Rb" value="0.124"/>
<Constant name="qa" value="7.2"/>
<Constant name="mmin" value="0.02"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v, tha,Ra,qa);
double b = trap0(-v, -tha,Rb,qa);
mtau = 1/(a+b)/qt;
if (mtau<mmin) mtau=mmin;
minf = a/(a+b);
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="htau" infvar="hinf">
<Constant name="q10" value="2"/>
<Constant name="thi1" value="-45"/>
<Constant name="thi2" value="-45"/>
<Constant name="thinf" value="-50"/>
<Constant name ="Rd" value="0.03"/>
<Constant name="Rg" value="0.03"/>
<Constant name="qd" value="1.5"/>
<Constant name="qg" value="1.5"/>
<Constant name="hmin" value="0.5"/>
<Constant name="qinf" value="4"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double a = trap0(v,thi1,Rd,qd);
double b = trap0(-v,-thi2,Rg,qg);
htau = 1/(a+b)/qt;
if (htau<hmin) htau=hmin;
hinf = 1/(1+Math.exp((v-thinf)/qinf));
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="stau" infvar="sinf">
<Constant name="q10" value="2"/>
<Constant name="vvh" value="-58"/>
<Constant name="vvs" value="2"/>
<Constant name ="a2" value="0.8" id="Na3-a2-semiconstant"/> <!-- a2 is the only change between soma, apical basal. -->
<Constant name="zetas" value="12"/>
<Constant name="gms" value="0.2"/>
<Constant name="vhalfs" value="-60"/>
<Constant name="a0s" value="0.0003"/>
<Constant name="smax" value="10"/>
<![CDATA[
double qt = Math.pow(q10, ((temperature-24)/10));
double c = 1 / (1+Math.exp((v-vvh)/vvs));
sinf = c+a2*(1-c);
double bets = Math.exp(1.e-3*zetas*gms*(v-vhalfs)*9.648e4/(8.315*(273.16+temperature)));
double alps = Math.exp(1.e-3*zetas*(v-vhalfs)*9.648e4/(8.315*(273.16+temperature)));
stau = bets / (a0s * (1 + alps));
if (stau<smax) stau=smax;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Na3-coded-apical.xml
<DerivedKSChannel id="Na3-coded-apical" from="Na3-coded-soma"> <ParameterChange to="Na3-a2-semiconstant" attribute="value" newText="0.5"/> </DerivedKSChannel>
Na3-coded-basal.xml
<DerivedKSChannel id="Na3-coded-basal" from="Na3-coded-soma"> <ParameterChange to="Na3-a2-semiconstant" attribute="value" newText="1"/> </DerivedKSChannel>
Naxfit-coded.xml
<KSChannel id="Naxfit-coded" permeantIon="Na" gSingle="30pS">
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="mtau" infvar="minf">
<Constant name="onebykt" value="0.039471195" info="AT 20 C!!"/>
<Constant name="z" value="4.82"/>
<Constant name="vHalf" value="-35.08" info="vhalf, mV"/>
<Constant name="gamma" value="0.49" info=""/>
<Constant name="tau" value="0.569" info=""/>
<Constant name="tmf" value="0.0304" info=""/>
<Constant name="tmr" value="0.1773" info=""/>
<![CDATA[
double rf = Math.exp (onebykt * gamma * z * (v - vHalf)) / tau;
double rr = Math.exp (-onebykt * (1-gamma) * z * (v - vHalf)) / tau;
double a = 1. / (1. / rf + tmf);
double b = 1. / (1. / rr + tmr);
mtau = 1 / (a + b);
minf = b * mtau;
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="htau" infvar="hinf">
<Constant name="onebykt" value="0.039471105" info="AT 20 C!!"/>
<Constant name="z" value="7.55"/>
<Constant name="vHalf" value="-46.4" info="vhalf, mV"/>
<Constant name="gamma" value="0.6" info=""/>
<Constant name="tau" value="11.4" info=""/>
<Constant name="tmf" value="0.564" info=""/>
<Constant name="tmr" value="1.42" info=""/>
<![CDATA[
double rf = Math.exp (onebykt * gamma * z * (v - vHalf)) / tau;
double rr = Math.exp (-onebykt * (1-gamma) * z * (v - vHalf)) / tau;
double a = 1. / (1. / rf + tmf);
double b = 1. / (1. / rr + tmr);
htau = 1 / (a + b);
hinf = b * htau;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Naxfit.xml
<KSChannel id="Naxfit" permeantIon="Na" gSingle="30pS">
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="c" to="o" z="4.82e" vHalf="-35.08mV" gamma="0.49" tau="0.569ms"
tauMinFwd="0.0304ms" tauMinRev="0.1773ms"/>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="o" to="c" z="7.55e" vHalf="-46.40mV" gamma="0.6"
tau="11.4ms" tauMinFwd="0.564ms" tauMinRev="1.42ms"/>
</KSComplex>
</KSChannel>
morph59.xml (truncated)
<morphml id="morph59">
<!-- XML file generated by NEURON 5.9 ModelViewer -->
<!-- Authors: Michael Hines and Sushil Kambampati -->
<!-- Yale University -->
<!-- Date: Fri Sep 7 12:52:05 BST 2007 -->
<cells>
<cell name="soma[0]">
<segments>
<segment id="1" name = "Seg0_soma[0]" cable = "0">
<proximal x="0" y="0" z="0" diameter="3.4"/>
<distal x="0.25" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="2" name = "Seg1_soma[0]" parent="1" cable = "0">
<distal x="0.5" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="671" name = "Seg0_basal[41]" parent="1" cable = "181">
<proximal x="0" y="0" z="0" diameter="1"/>
<distal x="-4.38737" y="1.85495" z="0" diameter="1"/>
</segment>
<segment id="672" name = "Seg1_basal[41]" parent="671" cable = "181">
<distal x="-8.77474" y="3.7099" z="0" diameter="1"/>
</segment>
<segment id="606" name = "Seg0_basal[23]" parent="1" cable = "163">
<proximal x="0" y="0" z="0" diameter="1"/>
<distal x="-5.95875" y="0.799243" z="0" diameter="1"/>
</segment>
<segment id="607" name = "Seg1_basal[23]" parent="606" cable = "163">
<distal x="-11.9175" y="1.59849" z="0" diameter="1"/>
</segment>
<segment id="593" name = "Seg0_basal[20]" parent="1" cable = "160">
<proximal x="0" y="0" z="0" diameter="0.4"/>
<distal x="-8.98517" y="-1.20517" z="0" diameter="0.4"/>
</segment>
<segment id="594" name = "Seg1_basal[20]" parent="593" cable = "160">
<distal x="-26.9555" y="-3.61552" z="0" diameter="0.4"/>
</segment>
<segment id="595" name = "Seg2_basal[20]" parent="594" cable = "160">
<distal x="-35.9407" y="-4.82069" z="0" diameter="0.4"/>
</segment>
<segment id="517" name = "Seg0_basal[0]" parent="1" cable = "140">
<proximal x="0" y="0" z="0" diameter="0.8"/>
<distal x="-1.33023" y="-0.562414" z="0" diameter="0.8"/>
</segment>
<segment id="518" name = "Seg1_basal[0]" parent="517" cable = "140">
<distal x="-2.66047" y="-1.12483" z="0" diameter="0.8"/>
</segment>
<segment id="3" name = "Seg0_soma[1]" parent="2" cable = "1">
<proximal x="0.5" y="0" z="0" diameter="3.4"/>
<distal x="0.55" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="4" name = "Seg1_soma[1]" parent="3" cable = "1">
<distal x="0.6" y="0" z="0" diameter="3.4"/>
</segment>
<segment id="736" name = "Seg0_basal[59]" parent="672" cable = "199">
<proximal x="-8.77474" y="3.7099" z="0" diameter="0.4"/>
<distal x="-15.8252" y="10.9693" z="0" diameter="0.4"/>
</segment>
<segment id="737" name = "Seg1_basal[59]" parent="736" cable = "199">
<distal x="-29.9261" y="25.4881" z="0" diameter="0.4"/>
</segment>
- and 2829 more lines -run1a.xml
<PSICSRun timeStep="0.05ms" runTime="120ms" startPotential="-65mV" morphology="morph59" environment="environment" properties="membrane" access="recording-a" stochThreshold="0"> <StructureDiscretization baseElementSize="10um"/> <info>Migliore et al CA1 pyramidal cell</info> <About> Model corresponding to figure 1a with a steady current injection at the soma and recording from two points on the apical dendrite, 200um and 400um from the soma. It shows some backpropagation to 200um but very little at 400um. </About> <About> This is not an exact reimplementation of the Migliore et al model but rather a nearby equivalent in PSICS using only the standard channel transition equation. Slow inactivation of the sodium channel has not been modelled as an independent gate that only partially closes (closing to 20% in the soma and to 50% in the dendrites) since the kinetics of this process do not appear to affect the model. Instead, the inactivated dendritic channels are simply left out of the model by providing two populations: one population, at 50% of maximum, that covers all active compartments and another, overlapping population, just for the soma. </About> <ViewConfig> <LineGraph width="500" height="400"> <XAxis min="0" max="250" label="time / ms"/> <YAxis min="-80" max="60" label="potential / mV"/> <LineSet file="psics-out-f.txt" color="green"/> <View id="whole" xmin="-10." xmax="120." ymin="-80." ymax="20."/> <View id="first" xmin="-2" xmax="20" ymin="-80." ymax="20."/> </LineGraph> </ViewConfig> </PSICSRun>
membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="150ohm_cm"
membraneCapacitance="1uF_per_cm2">
<PassiveProperties region="*apical*" membraneCapacitance="2uF_per_cm2"/>
<PassiveProperties region="*axon*" cytoplasmResistivity="50ohm_cm"/>
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<CellRegion match="*axon*">
<ChannelPopulation channel="Nax-coded" density="21.33per_um2"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="3.2per_um2"/>
</CellRegion>
<CellRegion match="*soma*">
<ChannelPopulation channel="Na3-coded-soma" density="10.67per_um2"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2"/>
</CellRegion>
<CellRegion match="*basal*">
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<ChannelPopulation channel="Na3-coded-basal" density="10.67per_um2"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2"/>
</CellRegion>
<CellRegion match="*apical*">
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<ChannelPopulation channel="Na3-coded-apical" density="10.67per_um2" distribution="fat500"/>
<ChannelPopulation channel="Kdr-coded" density="3.33per_um2" distribution="fat500"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2" distribution="kaprox"/>
<ChannelPopulation channel="Kadist-coded" density="16per_um2" distribution="kadist"/>
</CellRegion>
<DistributionRule id="fat500">
<RegionMask action="exclude" where="diameter .lt. 0.5"/>
<RegionMask action="exclude" where="path .gt. 500"/>
</DistributionRule>
<DistributionRule id="kaprox" expression="(1 + path / 100)">
<RegionMask action="exclude" where="path .gt. 100"/>
</DistributionRule>
<DistributionRule id="kadist" expression="(1 + path / 100)">
<RegionMask action="exclude" where="path .lt. 100"/>
<RegionMask action="exclude" where="path .gt. 500"/>
</DistributionRule>
<DensityAdjustment maintain="-65mV" vary="leak-Na, leak-K"/>
</CellProperties>
recording-a.xml
<Access id="recording-a"> <CurrentClamp at="4" hold="0.15nA"/> <VoltageRecorder at="Seg0_apical[12]"/> <VoltageRecorder at="Seg0_apical[27]"/> </Access>
Kadist-coded.xml
<KSChannel id="Kadist-coded" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<TauInfCodedTransition from="c" to="o" tauvar="ntau" infvar="ninf">
<Constant name="q10" value="5"/>
<Constant name="zetan" value="-1.8"/>
<Constant name="pw" value="-1" info=""/>
<Constant name="tq" value="-40" info=""/>
<Constant name="qq" value="5" info=""/>
<Constant name="vhalfn" value="-1" info="mV"/>
<Constant name="gmn" value="0.39"/>
<Constant name="nmin" value="0.2" info="minimum time constant"/>
<Constant name="a0n" value="0.1"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double zeta=zetan+pw/(1+Math.exp((v-tq)/qq));
double a = Math.exp(1.e-3*zeta*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ninf = 1/(1 + a);
double betn = Math.exp(1.e-3*zeta*gmn*(v-vhalfn)*9.648e4/(8.315*(273.16+temperature)));
ntau = betn /(qt*a0n*(1+a));
if (ntau < nmin) ntau = nmin;
]]>
</TauInfCodedTransition>
</KSComplex>
<KSComplex instances="1">
<OpenState id="o"/>
<ClosedState id="c"/>
<TauInfCodedTransition from="c" to="o" tauvar="ltau" infvar="linf">
<Constant name="q10" value="5"/>
<Constant name="zetal" value="3"/>
<Constant name="vhalfl" value="-56" info="mV"/>
<Constant name="qtl" value="1" info=""/>
<Constant name="lmin" value="2" info="minimum time constant"/>
<![CDATA[
double qt=Math.pow(q10, ((temperature-24)/10));
double a = Math.exp(1.e-3*zetal*(v-vhalfl)*9.648e4/(8.315*(273.16+temperature)));
linf = 1 / (1 + a);
ltau = 0.26 * (v + 50) / qtl;
if (ltau < lmin / qtl) ltau = lmin / qtl;
]]>
</TauInfCodedTransition>
</KSComplex>
</KSChannel>
Kdrca1.xml
<KSChannel id="Kdrca1" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="c" to="o" z="2.70e" vHalf="13.00mV" gamma="0.700" tau="50ms"
tauMinFwd="0.ms" tauMinRev="0.ms"/>
</KSComplex>
</KSChannel>
membrane-leak.xml
<CellProperties id="membrane-leak"
cytoplasmResistivity="150ohm_cm"
membraneCapacitance="1uF_per_cm2">
<PassiveProperties region="apical*" membraneCapacitance="2uF_per_cm2"/>
<PassiveProperties region="axon*" cytoplasmResistivity="150ohm_cm"/>
<ChannelPopulation channel="leak-Na" density="0.179per_um2"/>
<ChannelPopulation channel="leak-K" density="0.179per_um2"/>
<CellRegion match="*soma*">
<ChannelPopulation channel="Na3-coded-soma" density="21.33per_um2"/>
<ChannelPopulation channel="Kdrca1" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2"/>
</CellRegion>
<CellRegion match="*axon*">
<ChannelPopulation channel="Nax-coded" density="21.33per_um2"/>
<ChannelPopulation channel="Kdrca1" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="3.2per_um2"/>
</CellRegion>
<CellRegion match="*basal*">
<ChannelPopulation channel="Na3-coded-basal" density="21.33per_um2"/>
<ChannelPopulation channel="Kdrca1" density="3.33per_um2"/>
<ChannelPopulation channel="Kaprox-coded" density="16per_um2"/>
</CellRegion>
<CellRegion match="*apical*">
<ChannelPopulation channel="Na3-coded-apical" density="21.33per_um2" distribution="fat500"/>
<ChannelPopulation channel="Kdrca1" density="3.33per_um2" distribution="fat500"/>
<ChannelPopulation channel="Kaprox-code