p1-f3-1c-det (run1c.xml)
Migliore et al CA1 pyramidal cell
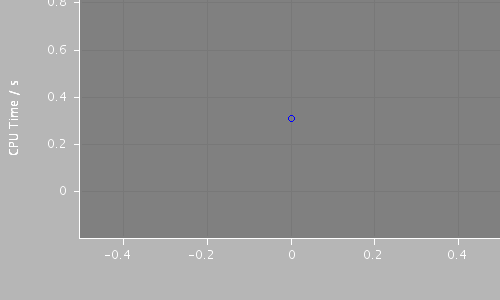
Total CPU time 0.3080 seconds; at 11:52:42 Fri 3 Aug 2007
| Compartments | Stochastic channels / cpmts | Continuous channels / cpmts | CPU Time / s |
| 325 | 0 / 0 | 883556 / 325 | 0.308 |
 |
Predefined views
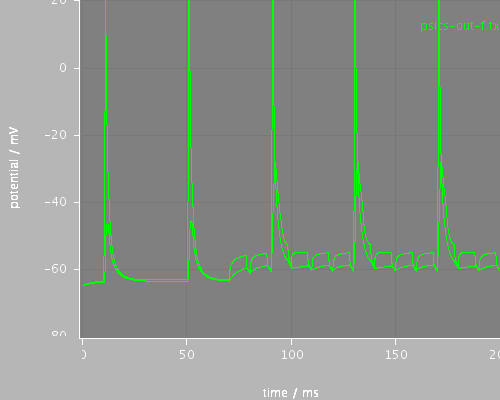
whole
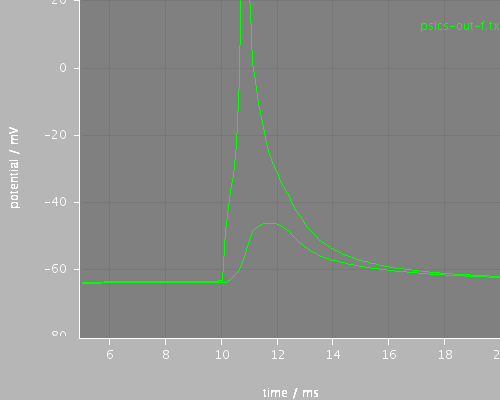
first
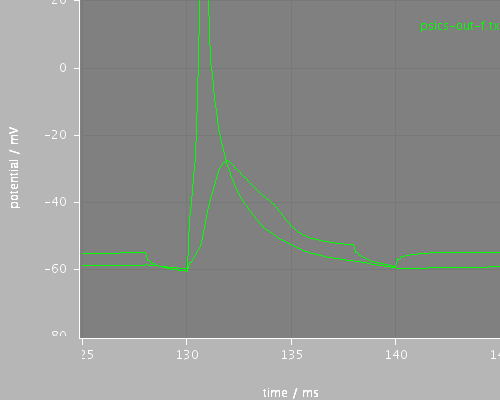
later
All files
| Model | Preprocessed | Outupt data | Reference data etc |
| run1c.xml morph.xml membrane.xml environment.xml leak.xml Nax.xml Kdrca1.xml Kaprox.xml Kadist.xml summary.xml view.xml recording-c.xml ns-leak.xml |
psics-out-fdata.txt |
psics-out-f.dat psics-out-f.txt |
Model
run1c.xml
<PSICSRun lib="." timeStep="0.1ms" runTime="200ms" startPotential="-65mV" morphology="morph" environment="environment" properties="membrane" access="recording-c" stochThreshold="0"> <StructureDiscretization baseElementSize="10um"/> <info>Migliore et al CA1 pyramidal cell</info> <ViewConfig> <LineGraph width="500" height="400"> <XAxis min="0" max="250" label="time / ms"/> <YAxis min="-80" max="60" label="potential / mV"/> <LineSet file="psics-out-f.txt" color="green"/> <View id="whole" xmin="-10." xmax="200." ymin="-80." ymax="20."/> <View id="first" xmin="5" xmax="20." ymin="-80." ymax="20."/> <View id="later" xmin="125" xmax="145." ymin="-80." ymax="20."/> </LineGraph> </ViewConfig> </PSICSRun>
morph.xml (truncated)
<!-- /tmp/cell.xml --> <!-- XML file generated by NEURON 5.6 ModelViewer --> <!-- Authors: Michael Hines and Sushil Kambampati --> <!-- Yale University --> <!-- Date: Tue Jul 31 11:15:50 BST 2007 --> <model> <morphology xmlns="http://morphml.org/namespace"> <setOfPoints> <point id="1" x="0" y="0" z="0" diam="3.4"/> <point id="2" x="0.25" y="0" z="0" diam="3.4"/> <point id="3" x="0.5" y="0" z="0" diam="3.4"/> <point id="4" x="0.5" y="0" z="0" diam="3.4"/> <point id="5" x="0.55" y="0" z="0" diam="3.4"/> <point id="6" x="0.6" y="0" z="0" diam="3.4"/> <point id="7" x="0.6" y="0" z="0" diam="5.8"/> <point id="8" x="0.892617" y="0" z="0" diam="5.8"/> <point id="9" x="1.18523" y="0" z="0" diam="5.8"/> <point id="10" x="1.18523" y="0" z="0" diam="7.4"/> <point id="11" x="2.36643" y="0" z="0" diam="7.4"/> <point id="12" x="3.54763" y="0" z="0" diam="7.4"/> <point id="13" x="3.54763" y="0" z="0" diam="8.4"/> <point id="14" x="4.06309" y="0" z="0" diam="8.4"/> <point id="15" x="4.57855" y="0" z="0" diam="8.4"/> <point id="16" x="4.57855" y="0" z="0" diam="9"/> <point id="17" x="7.12341" y="0" z="0" diam="9"/> <point id="18" x="9.66828" y="0" z="0" diam="9"/> <point id="19" x="9.66828" y="0" z="0" diam="8.4"/> <point id="20" x="10.749" y="0" z="0" diam="8.4"/> <point id="21" x="11.8297" y="0" z="0" diam="8.4"/> <point id="22" x="11.8297" y="0" z="0" diam="7.4"/> <point id="23" x="12.5256" y="0" z="0" diam="7.4"/> <point id="24" x="13.2215" y="0" z="0" diam="7.4"/> <point id="25" x="13.2215" y="0" z="0" diam="7"/> <point id="26" x="13.5837" y="0" z="0" diam="7"/> <point id="27" x="13.946" y="0" z="0" diam="7"/> <point id="28" x="13.946" y="0" z="0" diam="6.8"/> <point id="29" x="14.4567" y="0" z="0" diam="6.8"/> <point id="30" x="14.9674" y="0" z="0" diam="6.8"/> <point id="31" x="14.9674" y="0" z="0" diam="5.8"/> <point id="32" x="15.3851" y="0" z="0" diam="5.8"/> <point id="33" x="15.8028" y="0" z="0" diam="5.8"/> <point id="34" x="15.8028" y="0" z="0" diam="4.8"/> <point id="35" x="16.1278" y="0" z="0" diam="4.8"/> <point id="36" x="16.4528" y="0" z="0" diam="4.8"/> <point id="37" x="16.4528" y="0" z="0" diam="4.2"/> <point id="38" x="17.4266" y="0" z="0" diam="4.2"/> <point id="39" x="18.4003" y="0" z="0" diam="4.2"/> <point id="40" x="18.4003" y="0" z="0" diam="3.8"/> <point id="41" x="19.7579" y="0" z="0" diam="3.8"/> <point id="42" x="21.1156" y="0" z="0" diam="3.8"/> <point id="43" x="21.1156" y="0" z="0" diam="3.6"/> <point id="44" x="23.8164" y="0" z="0" diam="3.6"/> <point id="45" x="26.5173" y="0" z="0" diam="3.6"/> <point id="46" x="26.5173" y="0" z="0" diam="3"/> <point id="47" x="28.4558" y="0" z="0" diam="3"/> <point id="48" x="30.3943" y="0" z="0" diam="3"/> <point id="49" x="30.3943" y="0" z="0" diam="3"/> <point id="50" x="32.3851" y="0.841732" z="0" diam="3"/> <point id="51" x="34.376" y="1.68346" z="0" diam="3"/> - and 2082 more lines -
membrane.xml
<CellProperties id="membrane"
cytoplasmResistivity="150ohm_cm"
membraneCapacitance="1uF_per_cm2">
<ChannelPopulation channel="leak" density="0.00per_um2"/>
<ChannelPopulation channel="Nax" density="7per_um2" distribution="main"/>
<ChannelPopulation channel="Nax" density="12per_um2" distribution="soma"/>
<ChannelPopulation channel="Kdrca1" density="3.3per_um2"/>
<ChannelPopulation channel="Kaprox" density="16per_um2" distribution="prox"/>
<ChannelPopulation channel="Kadist" density="16per_um2" distribution="dist"/>
<ChannelPopulation channel="ns-leak" density="0.006per_um2" distribution="prox"/>
<ChannelPopulation channel="ns-leak" density="0.006per_um2" distribution="dist"/>
<DistributionRule id="main">
<RegionMask action="include" where="diameter .gt. 0.5"/>
</DistributionRule>
<DistributionRule id="soma">
<RegionMask action="include" where="path .lt. 40"/>
</DistributionRule>
<DistributionRule id="prox" expression="(1 + path / 100)">
<RegionMask action="exclude" where="path .gt. 100"/>
</DistributionRule>
<DistributionRule id="dist" expression="(1 + path / 100)">
<RegionMask action="exclude" where="path .lt. 100"/>
<RegionMask action="exclude" where="path .gt. 600"/>
</DistributionRule>
</CellProperties>
environment.xml
<CellEnvironment id="environment">
<Ion id="NSCAT" name="Non-specific cationic leak" reversalPotential="-40mV"/>
<Ion id="LEAK" name="Non-specific leak" reversalPotential="-65mV"/>
<Ion id="K" name="Potassium" reversalPotential="-90mV"/>
<Ion id="Na" name="Sodium" reversalPotential="55mV"/>
</CellEnvironment>
leak.xml
<KSChannel id="leak" gSingle="30pS" permeantIon="LEAK"> <OpenState id="o1"/> </KSChannel>
Nax.xml
<KSChannel id="Nax" permeantIon="Na" gSingle="30pS">
<KSComplex instances="3">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="c" to="o" z="4.82e" vHalf="-35.08mV" gamma="0.49" tau="0.569ms"
tauMinFwd="0.0304ms" tauMinRev="0.1773ms"/>
</KSComplex>
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="o" to="c" z="7.55e" vHalf="-46.40mV" gamma="0.6"
tau="11.4ms" tauMinFwd="0.564ms" tauMinRev="1.42ms"/>
</KSComplex>
</KSChannel>
Kdrca1.xml
<KSChannel id="Kdrca1" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="c" to="o" z="2.70e" vHalf="13.00mV" gamma="0.700" tau="50ms"
tauMinFwd="0.ms" tauMinRev="0.ms"/>
</KSComplex>
</KSChannel>
Kaprox.xml
<KSChannel id="Kaprox" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="c" to="o" z="3.42e" vHalf="0.15mV" gamma="0.655" tau="2.52ms"
tauMinFwd="1.58ms" tauMinRev="0.0ms"/>
</KSComplex>
<KSComplex instances="1">
<OpenState id="o"/>
<ClosedState id="c"/>
<VHalfTransition from="o" to="c" z="5.64e" vHalf="-63.64mV" gamma="0.443"
tau="6.17ms" tauMinFwd="3.17ms" tauMinRev="1.8ms"/>
</KSComplex>
</KSChannel>
Kadist.xml
<KSChannel id="Kadist" permeantIon="K" gSingle="30pS">
<KSComplex instances="1">
<ClosedState id="c"/>
<OpenState id="o"/>
<VHalfTransition from="c" to="o" z="4.47e" vHalf="-13.43mV" gamma="0.673" tau="0.825ms"
tauMinFwd="0.819ms" tauMinRev="0.153ms"/>
</KSComplex>
<KSComplex instances="1">
<OpenState id="o"/>
<ClosedState id="c"/>
<VHalfTransition from="o" to="c" z="5.64e" vHalf="-63.64mV" gamma="0.443" tau="3.17ms"
tauMinFwd="3.17ms" tauMinRev="1.8ms"/>
</KSComplex>
</KSChannel>
recording-c.xml
<Access id="recording-c"> <CurrentClamp at="2" hold="0nA"> <CurrentPulse start="10ms" duration="1ms" to="2nA"/> <CurrentPulse start="50ms" duration="1ms" to="2nA"/> <CurrentPulse start="90ms" duration="1ms" to="2nA"/> <CurrentPulse start="130ms" duration="1ms" to="2nA"/> <CurrentPulse start="170ms" duration="1ms" to="2nA"/> </CurrentClamp> <ConductanceClamp at="81" potential="0mV"> <ConductancePulse start="70ms" duration="8ms" to="3nS" repeatAfter="10ms"/> </ConductanceClamp> <VoltageRecorder at="80"/> </Access>
ns-leak.xml
<KSChannel id="ns-leak" gSingle="30pS" permeantIon="NSCAT"> <OpenState id="o1"/> </KSChannel>